Series information
During the winter-spring of 2022, I am enrolled in the genetic genealogy course, Research Like a Pro with DNA Study Group, from the team at Family Locket of Diana Elder, Nicole Dyer, and Robin Wirthlin. A weekly reflection journal is one course component. I’m sharing mine.
This week’s assignment: analyze your pedigree and list 2-3 brick walls that could be overcome using DNA; assess your closest DNA matches on each DNA testing website; and, perform basic clustering for closest matches.
Analyze Your Pedigree
I have two possible research objectives I am considering for my focus in this course, one modest, the other perhaps too ambitious. Both possible objectives are to confirm or refute the parentage of an ancestor using DNA analysis and documentary evidence. The more ambitious case dates to the late 1700s, involving my 3rd great-grandfather, Isaac Little (1799 — 1884, Ashe, NC) and determining Isaac’s father from among two brothers (Edmund Little or Peter Little) or perhaps their father, Charles Little (a 111-marker Y-DNA test strongly suggests that Isaac and this researcher are descendants of Charles’s ancestor, Abraham Little [1677, England — 1724, Virginia], mentioned above as my surname immigrant ancestor).
The more modest case is a non-paternity event (“NPE”) dating to the Civil War when a soldier, William McMillan (1830 — 1865, Ashe, NC), on leave in 1862, is said to have returned home not to his wife and children, but rather to have fathered my 2nd great-grandfather (James “Bawly” Bower, 1863 — 1960, Ashe, NC) with another woman, not his wife, my 3rd great-grandmother, Riley Bower (1840 — 1915, Ashe, NC). No shaming or judgment is implied or should be inferred; sometimes the most interesting stories arise out of challenging cases (just wait till you hear about Absolom Bower, scoundrel extraordinaire), not to mention that this researcher and a not-insignificant portion of the population of Ashe would never have been born but for this relationship.
Part 1: Basic Clustering / Leeds Method
Further below is an image depicting the results with which I finished the process of clustering with the Leeds Method. The steps to arrive here are described below, but I wanted to start with a glimpse of the final Leeds chart so that one could have a sense toward where we are headed. First, though, I need to note some background, identify a problem, and suggest a solution.
Background: All 32 of my 3rd great-grandparents settled into one Appalachian county by 1820. My sixty most-recent ancestors were born, lived, and died there, in Ashe County, North Carolina. My parents left Ashe County in the mid-1960s, the first of my direct ancestors to leave Ashe in 140 years; sometimes they say they escaped (left of their own free will) and at other times they will say that they were exiled (“asked” to leave); both are in jest.
Problem: On 20 August 2020, Dana Leeds wrote: “The Leeds Method, unfortunately, does not work well in the following situations: You have endogamy or pedigree collapse in your family tree.” This is certainly the case in my family and community. The vast majority of my matches above 90 cM share multiple relationships with each other. And this problem is not unique to my family. I manage DNA kits for more than a dozen people with ties to Ashe County, and, not only do test-takers of Ashe County share multiple relationships, but most of their significant matches (above 90 cM) also share multiple relationships with each other. This problem was made visibility clear to me in 2018 when the administrator of the Facebook group “Roots of Ashe County, NC Families Genealogy & DNA Research” shared a DNA matrix comparison of some of the members of the group with each other. Of a group of 300 members with ties to Ashe County, almost all had significant matches with multiple other group members, previously unknown to each other (that is, not friends or family with known relationships). The way that these multiple relationships and pedigree collapses occur can be illustrated with one of many possible examples in my own family; in my personal case, the most recent instance of pedigree collapse is the relationship between my paternal grandfather and my paternal grandmother:
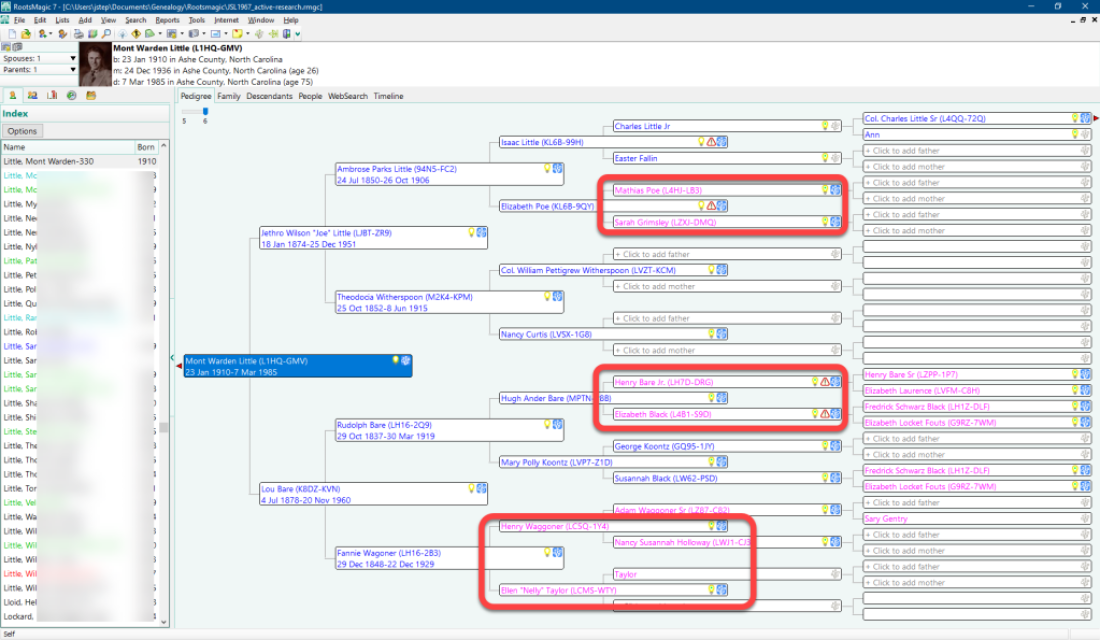
As seen in the image of my grandfather’s pedigree chart above, where my paternal grandfather’s ancestors are colored blue and my paternal grandmother’s ancestor are colored fuchsia (light purple), my grandfather shares one set of great-grandparents and two sets of second great-grandparents with my grandmother. And again, this is hardly the only instance of pedigree collapse in my family, and this is common throughout the heritage of Ashe County. So doing genetic genealogy in Ashe County is messy, but, to repurpose a phrase about New York City, if you can do genetic genealogy in Ashe County, you can do it anywhere. It’s just harder, and you have to be more careful.
My Hypothesis: I want to test an idea: That the Leeds Method can still be useful in families and communities with significant pedigree collapse and endogamy, but with modifications to the method, namely, that more matches must be searched and screened, looking for matches without significant overlap. Before stepping through the modified Leeds process I used, here is a glimpse of the final chart with which I ended. This is still a work-in-progress, but I finished this experiment with a Leeds chart with fairly well-defined clusters. Further analysis is needed and to evaluate the usefulness of this modified Leeds Method. But it appears to be a promising start. I am seeking feedback on the validity of this modified Leeds Method for dealing with endogamy.
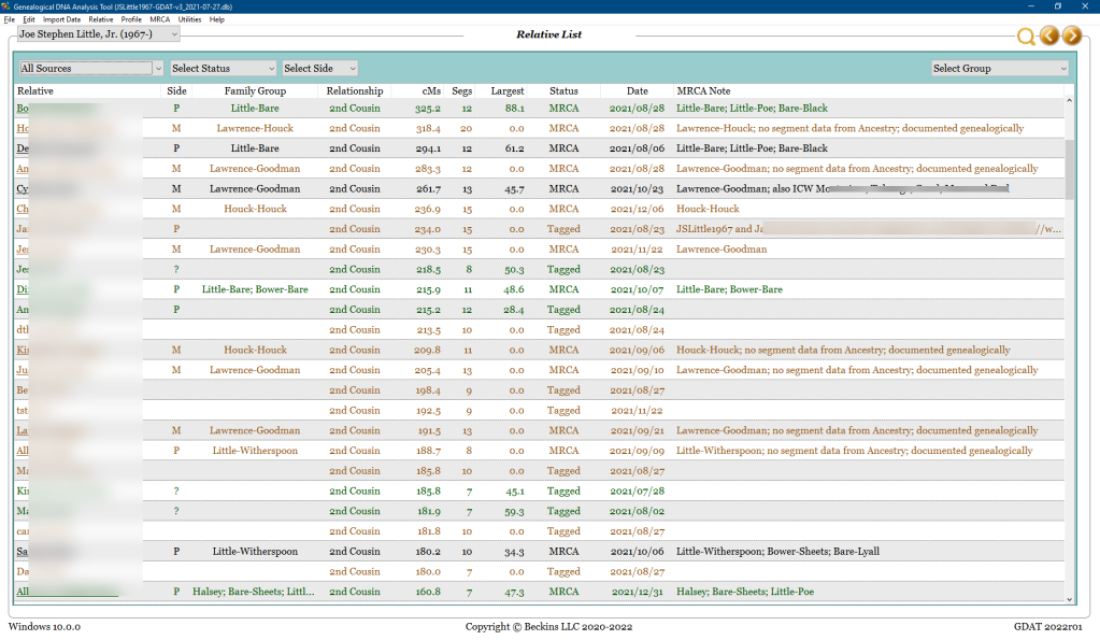
These are the steps I took to get the modified Leeds Method chart above. I have had my autosomal DNA tested at Ancestry, 23 and Me, My Heritage, and FamilyTreeDNA, and the results of all those tests have been uploaded to GEDmatch. Both my father and my mother have also tested at Ancestry, and I have uploaded those results also to GEDmatch, and shared them at My Heritage and FamilyTreeDNA. Since July 2021, I have been using the database application Genealogical DNA Analysis Tool (“GDAT”) to store and analyze this genetic genealogy information concerning genetic relatives, match data, segment data, and determination of MRCA-couples. I use the data gathering tool DNAGedcom to collect relative, match, and segment data from each of the testing sites and GEDmatch, and that data is imported into GDAT. This data was most recently refreshed during the second week of February 2022.
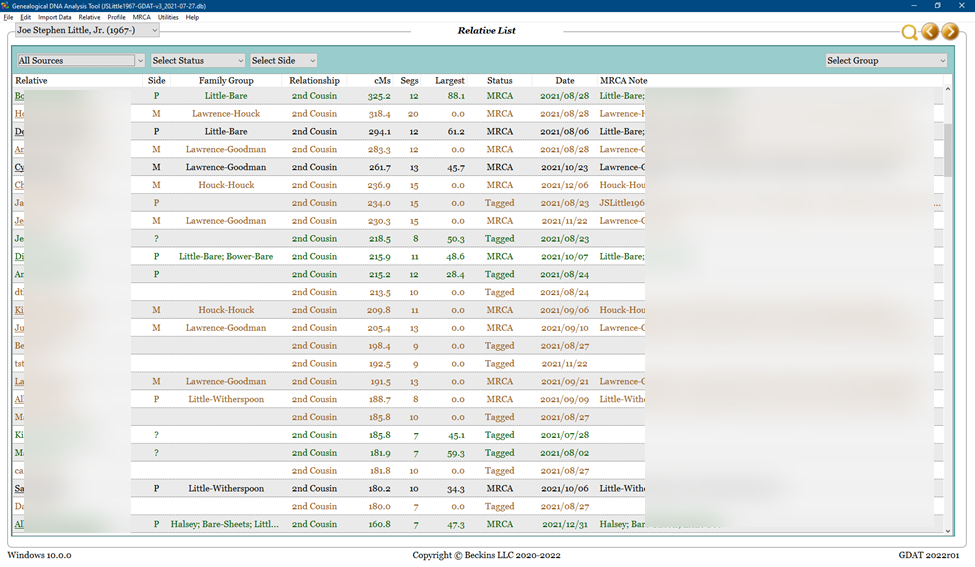
The relative, match, and segment data in GDAT can be viewed in several ways. Here is a view of my top matches above 75 cM:
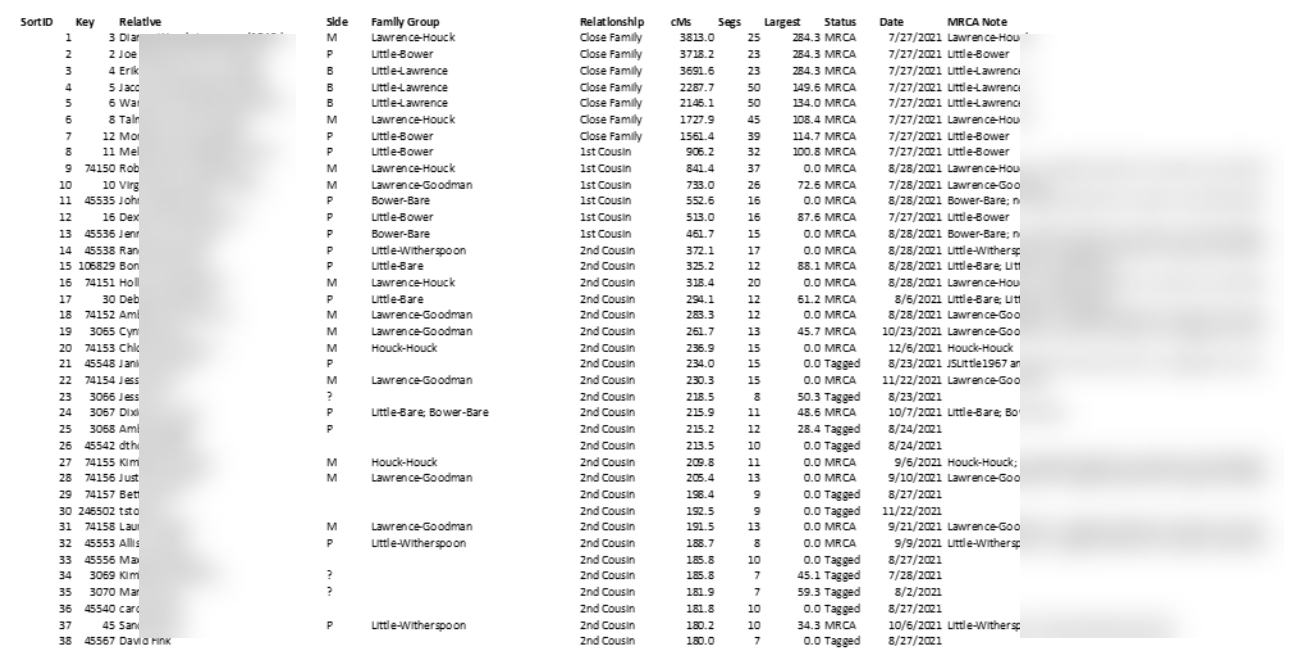
Data in GDAT can be exported in various formats, for example, as an Excel file, a CSV file, or a generic, open-source SQLite database file. Here can be seen CSV data from GDAT as imported into Excel:
Knowing that many of my top matches would contain multiple relationships due to pedigree collapse and therefore less well-suited to Leeds Method clustering, I cast a wider net, looking further for matches without multiple relationships. To cast this wider net, I exported all of my matches between 400 cM and 75 cM. This initial group contained about 150 matches. The standard Leeds Method processing was applied to those 150 matches, yielding this first chart:
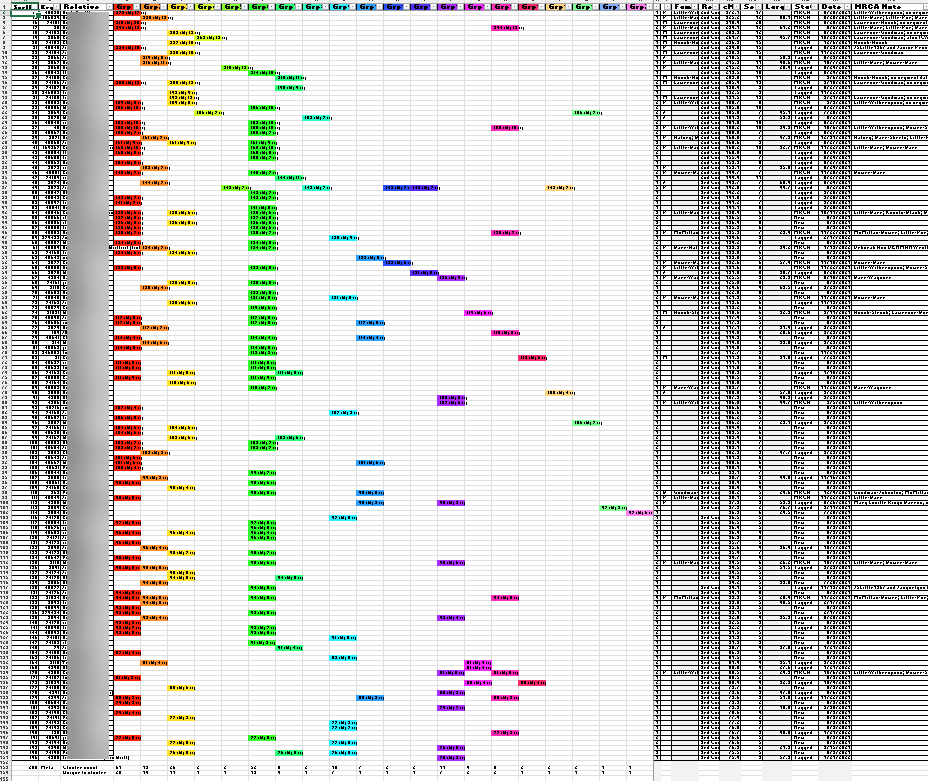
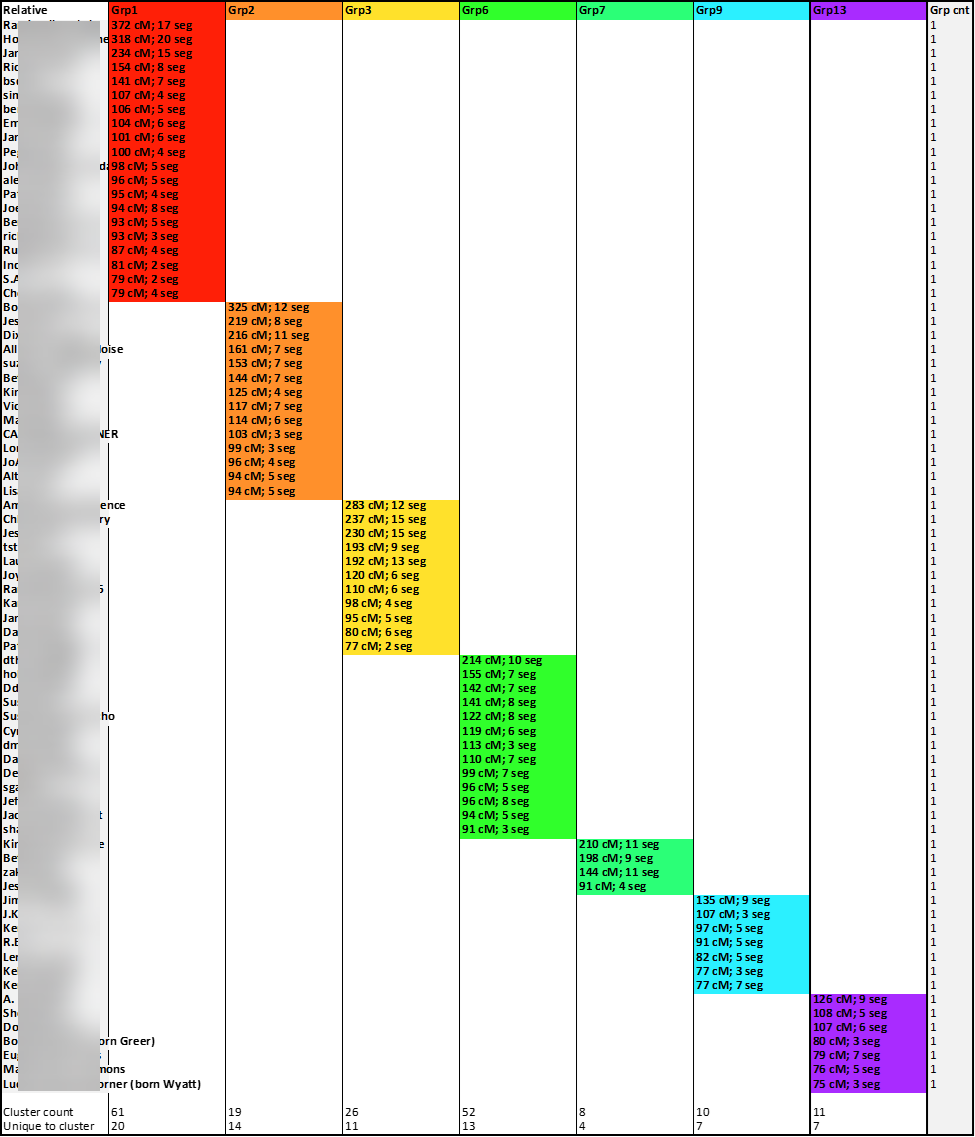
Instead of only coloring the cells of shared matches, I used the available match data to also include in the cell some match data, specifically, the total number of shared cM between the tester (me) and the match and the number of segments with the match. And, because those colored cells indicating matches also contained data, I was able to use the Excel COUNTA formula to determine how many cluster-groups each relative-match belonged. The column to the right of the last colored cluster-group (Group Count: “Grp cnt”) shows the sum of the number of cluster-groups to which a relative-match belongs. And as expected, there was evidence of significant overlap:

This highlight illustrates some of the higher instances of overlap: “Match A” matched five cluster-groups, “Match B” matched four cluster-groups, and nearly sixty of my matches overlaps two or three groups. These overlapping matches are the first batch of matches I will set-aside for my modified Leeds Method analysis.
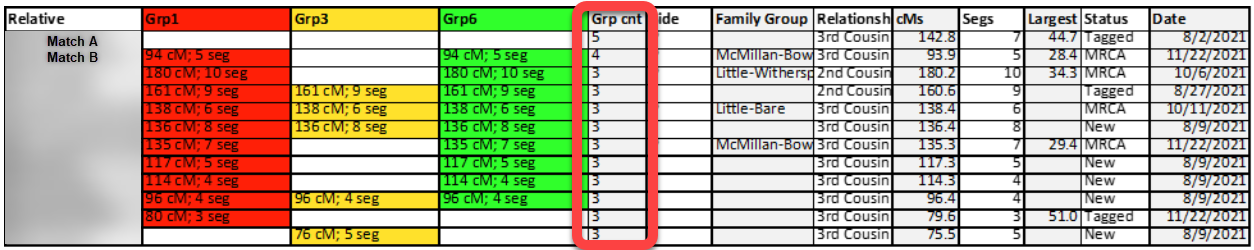
The second set of matches that were set-aside are cluster-groups that contained only one or two relative-matches. I used another COUNTA formula at the bottom of each cluster-group column to calculate how many relative-matches were included in each group, as seen here:

And so, I used the Excel method of “hiding” certain rows and columns. Rows containing relative matches belonging to two or more clusters were hidden. And cluster columns with only one or two members were hidden. This reduced the number of working relative-match down from 150 to 78 and the number of cluster-groups from 20 to 7. Each of the relative-matches show here share between 75 and 400 cM with the tester (me) and, despite the impactful and significant heritage of Appalachian pedigree collapse, because of the filtering and screening, each of the remaining 78 matches belongs to only one cluster-group:

A closer look at the results at this point is shown here:
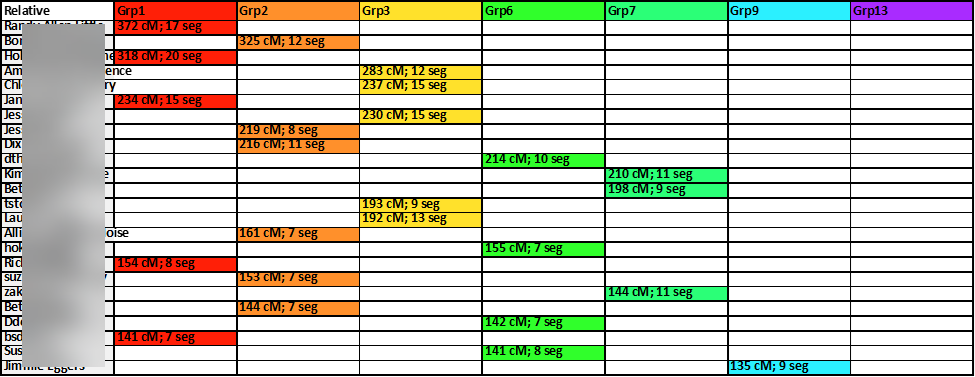
Then, sorting the columns by color, from first to last, the cluster-groups are gathered together:

A closer look is provided here:
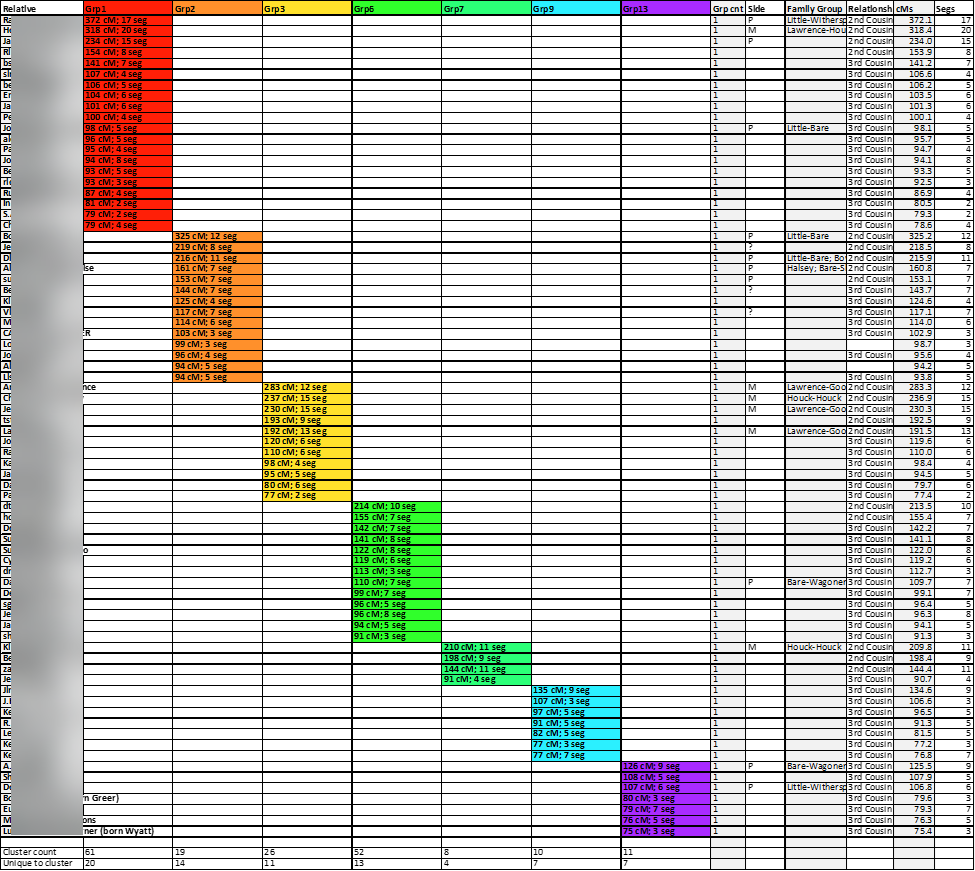
The removal of the row lines improves the appearance and readability of the chart:
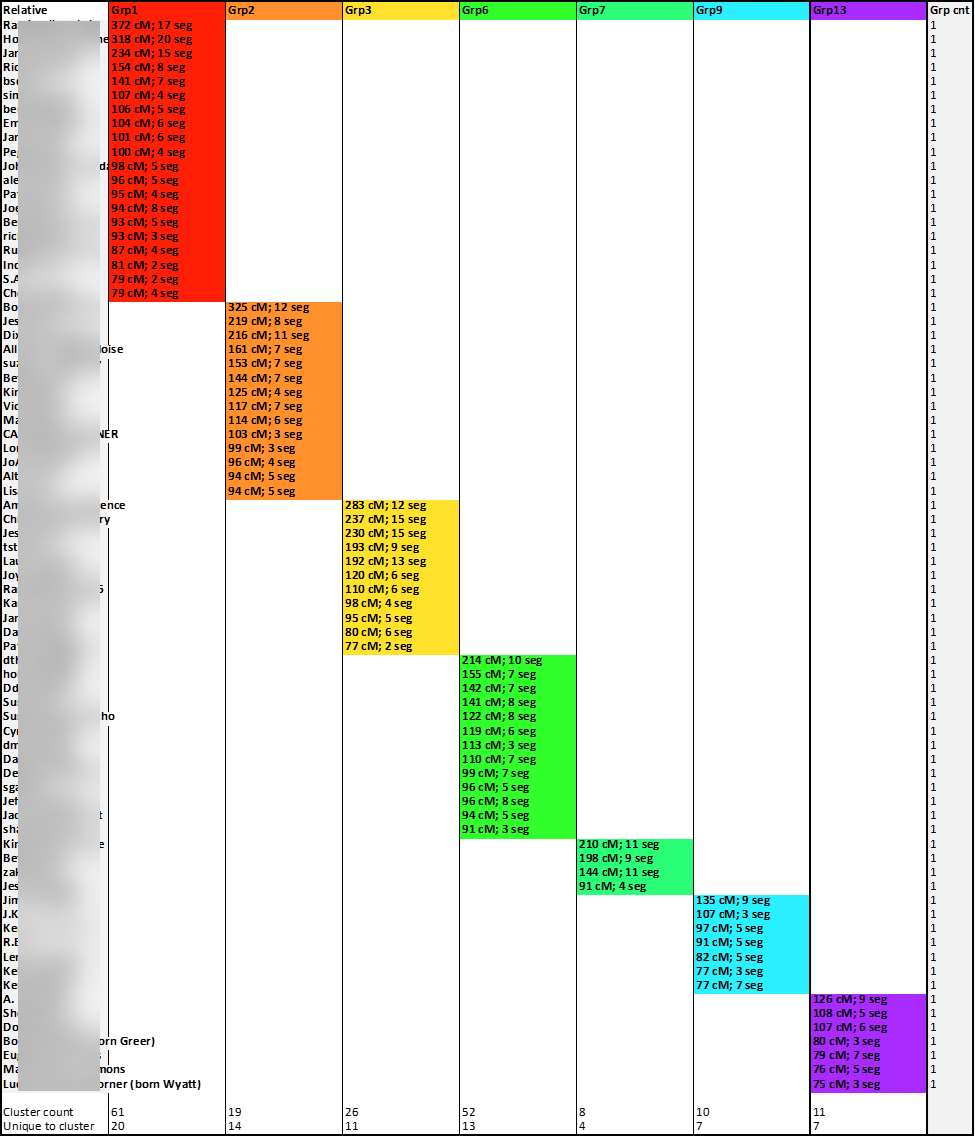
These cluster-groups composed of unique relative-matches should help me determine the most recent common ancestor couples among each group.
Next Steps: Considering my two possible research questions (the parentage of Isaac Little or the parentage of James “Bawly” Bower), the identification of the MRCA-couples of the seven cluster-groups found here may shed light on the most fruitful path of further research. These seven cluster-groups and the unique relative-matches highlighted here will be a focus in the coming weeks.
Later Possibilities: When the MRCA-couple for each cluster-group is determined, it may be possible to do further analysis with the relative matches that were set-aside earlier due to cluster-group overlap caused by pedigree collapse. At that point, it may be possible to identify the specific ancestors involved in the pedigree collapse. Further, a closer, detailed segment analysis of relative-matches may then determine which ancestor-couples contributed a specific DNA chromosome segment despite multiple relationships between the tester and the match, by using the unique relative-matches above with the known multiple relationships. That methodology will be worked out later.
Part 2, Assess Matches in Airtable
I have been excited about beginning to work with Airtable this week. Although perhaps dangerously amateurish, I have been using a relational database (though not Airtable) for about a year in my endeavor to become adept in genetic genealogy.
Since July 2022, I have been using Genealogical DNA Analysis Tool (“GDAT”) to store and analyze my DNA relative-matches. Personally, I have tested at Ancestry, 23 and Me, My Heritage, and FamilyTreeDNA, and have uploaded those results to GEDmatch. It soon became apparent that the duplication, triplication, and worse, of effort to work with match results and segment analysis at four or five testings sites and GEDmatch may not be the most efficient use of time and energy. So while the online genealogical sites may excel, each in their own way, at some aspects of genetic genealogy (e.g., Ancestry for records research, My Heritage for DNA work, etc.), none alone are a great place to hold all your data and work across multiple testing sites. So I use RootsMagic to store my genealogical information. And, as I mentioned, for about a year I have been using GDAT to hold and analyze all my DNA relationship, match, and segment data from all the testing sites in one application. The most significant drawbacks to GDAT, however, are a severe learning curve, terse documentation, and a smallish community of users to offer peer support. Like many things in life, however, I have found that with hard work comes great reward, and that has been my experience with GDAT. Learning to use the tool has taught me much about genetic genealogy and has yielded positive results, and the investment of time and energy really begins to pay dividends after a while. The screenshot above shows that currently in my GDAT database are more than a quarter million DNA relatives, 300,000 DNA segments and triangulations, and nearly two million in-common-with relationships among matches. These statistics are for the 15 DNA kits for which I use GDAT to hold and analyze relationship, matches, and segment data.
I have heard mention of Airtable before, but have not used it previously. The much larger user-base, better documentation, ease-of-use, and cloud-based always-accessible online access is appealing. For these and other reasons, instead of merely copying and populating Nicole’s sample database with my own hand-entered information, I am attempting to recreate from scratch Nicole’s database (so that I can better know how to build one) and to import some of the information currently in my GDAT database into this new Airtable base. In the image above, you can see how far I was able to get this week (a full-size image is included at the end of this journal entry).
These are the steps I used to re-create Nicole’s base and to populate it with my GDAT data. This image depicts a view of some of my top matches in GDAT. Most of the columns are self-explanatory; the different colors of matches indicate the original testing company, and the underlined names indicate that a family tree is available for viewing.
One of the many invaluable features of GDAT is the ability to export your data in several formats. The image here shows how I started this process by exporting my data out of GDAT into a generic open-source SQLite database. This generic database export can then be opened in any number of database programs such as MS Access, SQLite Expert, or the free dBeaver; this is the same concept as using MS Word or Google Docs to open a text file, or using MS Excel for Google Sheets to open a CSV file. The database that GDAT exports has all the information that I have been collecting for years from Ancestry, 23 and Me, My Heritage, FamilyTreeDNA, and GEDmatch; the export file also preserves the relationships and analysis that has been recorded during your earlier work sessions.
For example, this image shows the tables of data that GDAT has exported and a small sample of segment match data, as seen in a database management application such as SQLite Expert:
One great power of a relational database such as Airtable, SQLite, or GDAT is the ability to sort, filter, and combine tables of information. When desired, however, the raw data can be exported into the simplest of formats, a plain text file, such as a CSV file. Here, for example, is a glimpse of a CSV file as seen in Excel before uploading to Airtable:
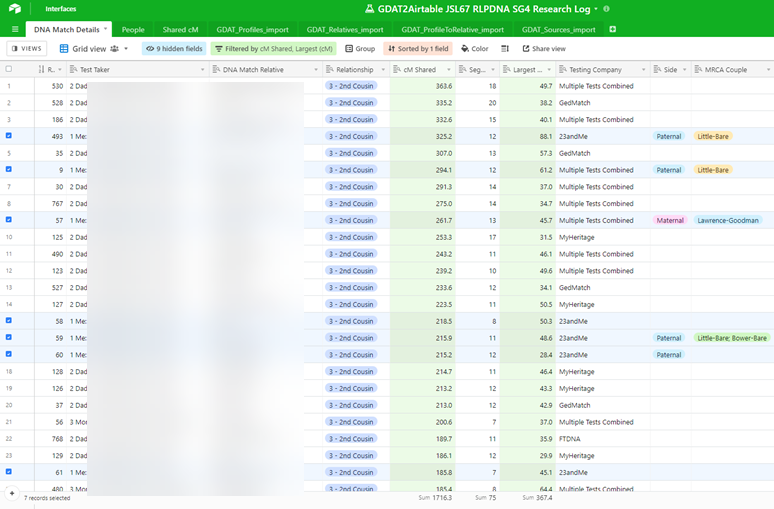
After uploading as a new imported table, the same data is now in Airtable, ready to be sorted, filtered, and combined with other information in order to analyze possible genetic relationships to answer genealogical questions, such as “Who is the father of James Eli ‘Bawly’ Bower?”
Here, I have begun to combine information from different tables to re-create Nicole’s “DNA Match Details” table. Using this export-from-GDAT and import-into-Airtable method, I was able in fairly short order to populate my Airtable base with the top 640 of my DNA relatives from Ancestry, 23 and Me, My Heritage, FamilyTreeDNA, and GEDmatch with whom I share 75 cM or greater of DNA, along with the 795 in-common-with relationship between those matches and either me, my father, or my mother, which Airtable quickly determined and revealed as 148 distinct DNA matches between those relatives and me, my father, and my mother, as shown in this closing view:
Some may ask, Why go to the trouble? Isn’t exporting the data from GDAT and re-creating the database in Airtable a bit like dismantling a Porsche and reassembling it as a Volkswagen on the other side of the garage? Maybe. After all, GDAT is a custom-built application, specifically designed for genetic genealogy, with its own genetic analytical tools and genealogical abilities. But it’s not an either/or situation. Airtable might be a useful way to collaborate. And, Airtable might be an easier way to integrate information from several databases. For example, RootsMagic, where many genealogists store their family tree and family history data, is also a glorified SQLite database. The genealogical information in a RootsMagic database can also be exported to a generic format. And so, perhaps Airtable might be the place where one can integrate genetic information from GDAT and the genealogical data from RootsMagic. And pulling together and leveraging this information from separate repositories might more easily help answer genealogical questions and solve family mysteries, allowing forgotten stories to be retold.
2 thoughts on “Reflection Journal: Week 1: Assess Your DNA Matches & Analyze Your Pedigree”